COMPUTATIONAL GENOMICS
THE "HOW" OF SCIENCE

METHODS OF DNA
SEQUENCING
WHAT IS DNA SEQUENCING??
DNA sequencing is the process of identifying the order of the four bases that make up DNA. The specific order of the sequence determines what kind of genetic information the DNA codes for. Scientists sequence DNA to determine which parts of the DNA segment contain genes that code for the synthesis of proteins and what parts of the DNA are regulatory. Sequencing DNA can also identify which mutations exist in the genome that could correlate to a specific disease.

HOW DO YOU SEQUENCE DNA?
MAXAM-GILBERT SEQUENCING TECHNIQUE
The first method for sequencing DNA was developed by Allan Maxam and Walter Gilbert in 1976–1977 and is subsequently called Maxam Gilbert Sequencing. This technique is widely considered one of the pioneers that led the way for a lot of the next generation sequencing techniques that would be developed in coming years.
This method was time-consuming and was very manual which could lead to errors. It could only sequence about 200-300 bases every few days. The process also required large amounts of radioactive material and chemicals. With the development of next-generation sequencing, Maxam-Gilbert sequence is not regularly used [1].

Diagram above shows the steps of sequencing DNA using the Maxam-Gilbert technique [1]
WHAT ARE THE STEPS OF MAXAM-GILBERT SEQUENCING METHOD?
1) DNA is denatured into a single-strand and labeled on the 5' end.
2) The DNA is selectively cut using chemicals that can differentiate between the 4 bases and cleave accordingly
3) The DNA is loaded onto a polyacrylamide gel to separate the cut pieces according to size. A radioactive tag is used so that the DNA can be seen
4) The sequence of bases is read in the gel by deciphering how the DNA strands were cut by the selective chemicals back in step 2 [1].
POLONY SEQUENCING








The Polony sequencing method, developed in the laboratory of George M. Church at Harvard, was among the first high-throughput sequencing systems and was used to sequence a full E. coli genome in 2005 [2].
WHAT ARE THE STEPS OF THE POLONY SEQUENCING METHOD?
1) Genomic DNA is cut apart by restriction enzymes.
2) DNA is repaired. The 5' end is repaired with a blunt end that has a phosphate group attached, allowing it to bind with adapter oligonucleotides in a future step. An A is added to the 3' end of the sheared DNA, giving it a sticky end. After being repaired, the DNA is loaded into a gel.
3) An oligonucleotide with two recognition sites binds with the sticky and blunt end of the DNA so that the DNA strand is now a circle.
4) The now-circularized DNA goes through rolling circle replication, which is a type of replication that generates several new copies of circular DNA.
The new copies are now digested by a new type of restriction enzyme that cuts a few base pairs out of the sequence. The DNA molecule is no longer in a circular shape.
5) The DNA is repaired again and primers are added on either end to get ready for emulsion PCR
6) Emulsion PCR is used to amplify the DNA. This process uses a bead inside a water droplet embedded in a solution of oil.
7) A coverslip for a slide is treated with aminosilane. The beads from the ePCR are mixed with a gel called acrylamide and poured onto a microscope slide and the coverslip is placed on top. The beads can now bind to the coating of aminosilane and spread out on the slide.
8) The DNA is marked with a fluorescent marker and an oligonucleotide is used to sequence DNA [2].
This diagram shows the various steps that are used during Polony Sequencing [2].
454 PYROSEQUENCING
454 Pyrosequencing is a type of pyrosequencing that focuses on measuring the amounts of pyrophosphates that are released during DNA synthesis. Pyrosequencing was first conceptualized in 1993 by Bertil Pettersson, Mathias Uhlen, and Pål Nyren [3].
WHAT ARE THE STEPS OF 454 PYROSEQUENCING?
-
A sample of DNA is obtained.
-
Restriction enzymes are used to specifically cut the DNA into several 400-600bp fragments.
-
Adaptors sequences are attached to the DNA.
-
Beads of resin are mixed with the DNA sample.
-
The DNA in the beads is complementary to the DNA in the sample. The DNA fragments bind to the beads.
-
During the binding process between the DNA and the beads, the bond that keeps the two original strands of DNA is broken, causing the DNA to become single stranded.
-
PCR is used to replicate the single-stranded DNA.
-
The beads undergo a filtration process to eliminate any errors in the sample.
-
-
The beads are placed on a sequencing plate along with separate enzyme beads containing a primer and DNA polymerase.
This video shows some of the procedures used during Pyrosequencing [1].

This image shows a magnified view of a bead landing on the crevice in the sequencing plate [3].

This diagram shows the various steps that are used during 454 Pyrosequencing [4].
-
The new enzyme beads bind to the DNA from the original sample.
-
A wave of Adenine, Cytosine, Guanine,
-
and Thymine are introduced to the wells in that order.
-
A light is given out, and the results are recorded.
-
Scientists are able to tell the sequences of DNA based on the intensity of light.
-
Scientists plot the resulting intensities and are able to figure out the specific sequence of the DNA [4].
ILLUMINA SEQUENCING

This image shows a visual representation of the steps of illumina sequencing [5].

This image shows the final product of the sequencing that is ready to analyzed [6].
Illumina Next Generation Sequencing Technology is an efficient and accurate method for sequencing DNA. The method uses dye- terminators to identify the single bases that make up the DNA strands.
The Illumina method was developed by Shankar Balasubramanian and Klenerman at Cambridge University. They later founded Solexa, which is the company that now owns the rights to Illumina [5].
WHAT ARE THE STEPS OF THE ILLUMINA SEQUENCING TECHNOLOGY?
-
The DNA is cut into fragments with about 100-150 base pairs.
-
DNA adaptors are attached to the DNA sample.
-
Using sodium hydroxide, the DNA is converted into a single-stranded form.
-
The DNA is attached to a flowcell. The complementary DNA on the slide binds to the sample DNA.
-
The DNA is replicated using PCR.
This video shows some of the procedures used during Illumina Sequencing [2].
-
Nucleotide bases and DNA polymerase are added to the DNA. The segments now join across the flowcell to form "bridges" of double-stranded DNA on the surface of the slide.
-
The bridges are broken down back into single-stranded DNA using heat. This leaves clusters of identical DNA sequences stretched across the flowcell.
-
Primers and fluorescently-labeled nucleotide bases coding for the termination of DNA synthesis are added to the slide.
-
The DNA binds to the primer.
-
The DNA polymerase (added in step 6) binds to the primer and adds the fluorescent terminator base to the new strand of DNA. This terminator base acts as a cap that prevents any more bases to be added to this strand of DNA.
-
A laser activates the fluorescent color of the terminator. Each version of the terminator base (A, C, G, or T) gives off a different color. The specific color of the terminators is detected and recorded by a camera [5].
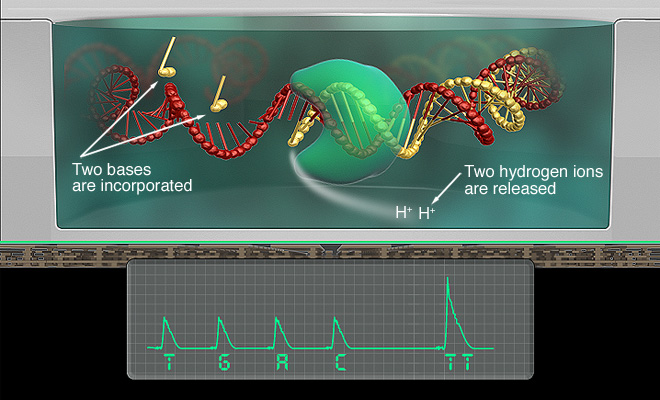
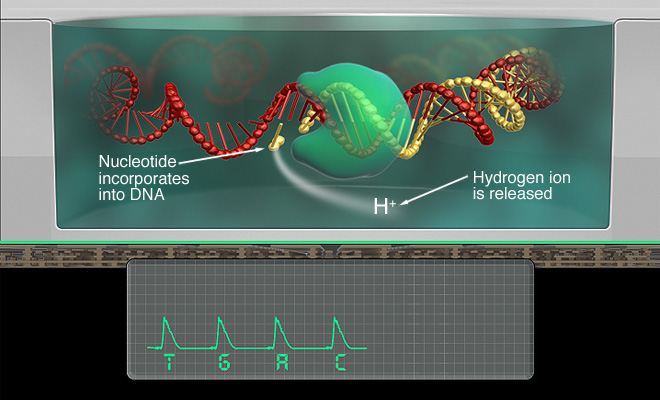
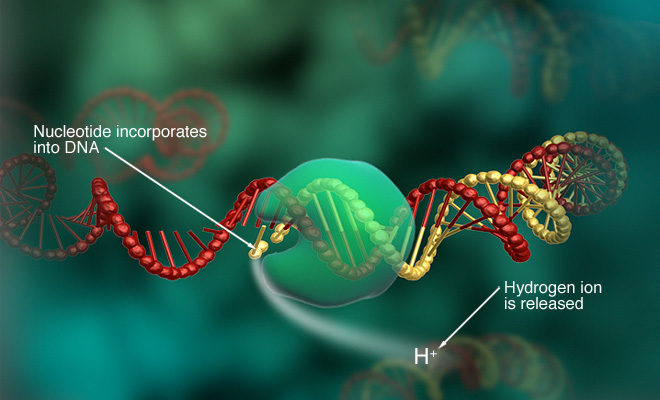
ION SEMI-CONDUCTOR TECHNOLOGY
Ion Semi-conductor technology was developed by Ion Torrent Systems Incorporated in 2010. It was licensed by DNA Electronics Ltd.
During the sequencing process, a strand of deoxyribonucleotide triphosphate (dNTP) is added to a microwell containing a sample of DNA. If the dNTP is complementary to the sample of DNA, the two strands will bond, causing the release of a hydrogen ion. This hydrogen ion will increase the ph in the solution.
This change in ph is detected and recorded by the sensor in the technology [6]

This video shows some of the procedures used during Ion Semi-Conductor Technology [3].
These images depict some of the nuances of ion semi-conductor technology and its applications [7].